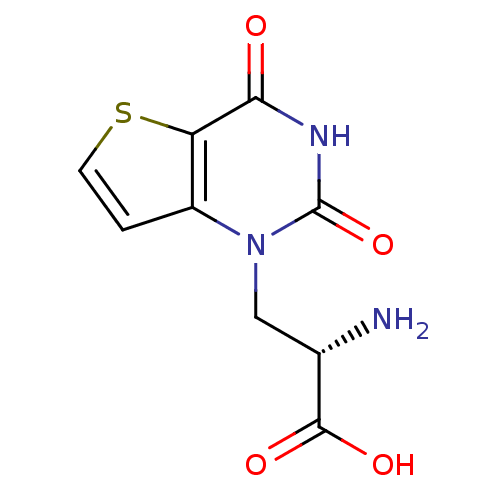
BDBM50252922 (S)-1-(2'-Amino-2'-carboxyethyl)thieno[3,2-d]pyrimidin-2,4-dione::CHEMBL492630
SMILES N[C@@H](Cn1c2ccsc2c(=O)[nH]c1=O)C(O)=O
InChI Key InChIKey=XTASTURWHDUENT-BYPYZUCNSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 50252922
Found 3 hits for monomerid = 50252922
Affinity DataKi: 112nMAssay Description:Displacement of [3H]AMPA from rat GluA2 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 112nMAssay Description:Displacement of (R,S)-[5-methyl-3H]AMPA from rat recombinant flop iGluR2(R) expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataEC50: 1.18E+4nMAssay Description:Agonist activity at cyclothiazide-desensitized rat recombinant flip iGluR2(Q) expressed in Xenopus laevis oocytes by two electrode voltage-clamp elec...More data for this Ligand-Target Pair